Publication
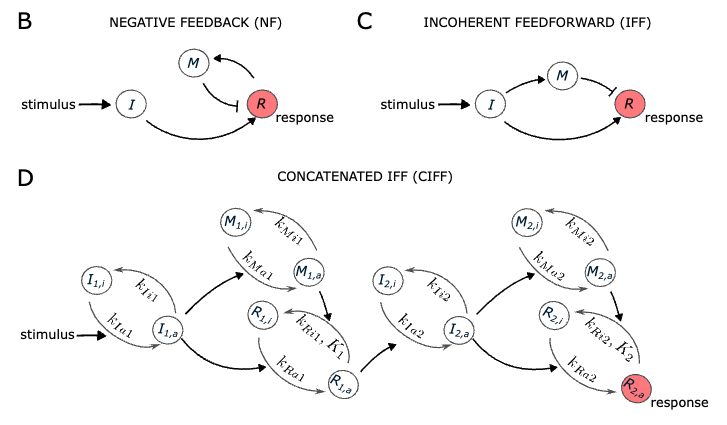
Eckert, L., Vidal, M. S., Zhao, Z., Garcia-Ojalvo, J., Martinez-Corral, R., & Gunawardena, J. (2024). Learning in single cells: biochemically-plausible models of habituation. bioRxiv, 2024-08.
TLDR: We show that simple molecular networks can exhibit all hallmarks of habituation. Our models suggest that individual cells may display learning-like behavior as rich as that found in multicellular organisms.
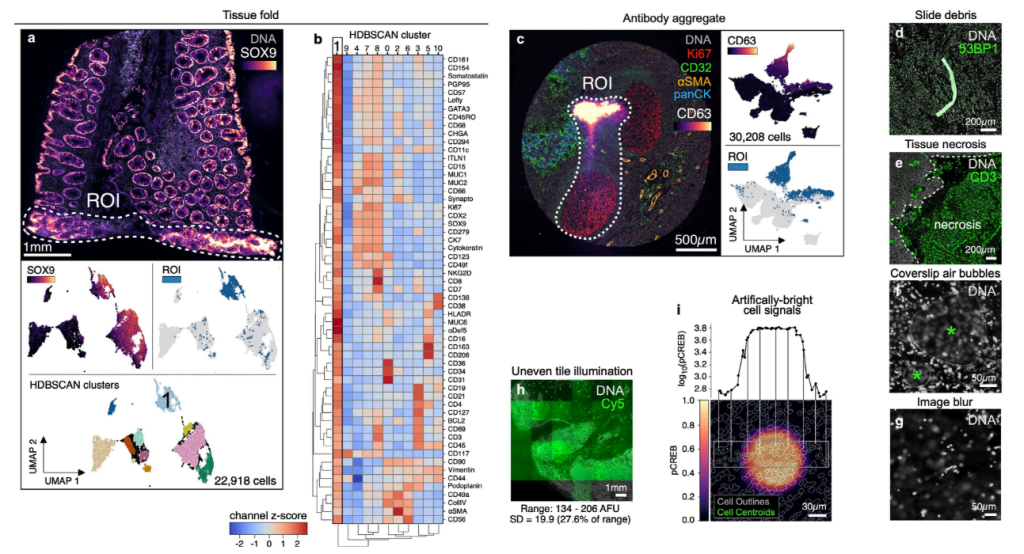
Baker, G. J., Novikov, E., Zhao, Z., Vallius, T., Davis, J. A., Lin, J. R., … & Sorger, P. K. (2023). Quality control for single cell analysis of high-plex tissue profiles using CyLinter. (Accepted to Nat. Method, yay)
TLDR: Image artifacts in data produced from high-plex spatial profiling methods can dramatically impact single-cell data analysis. We introduce CyLinter, an interactive tool, to address this problem.
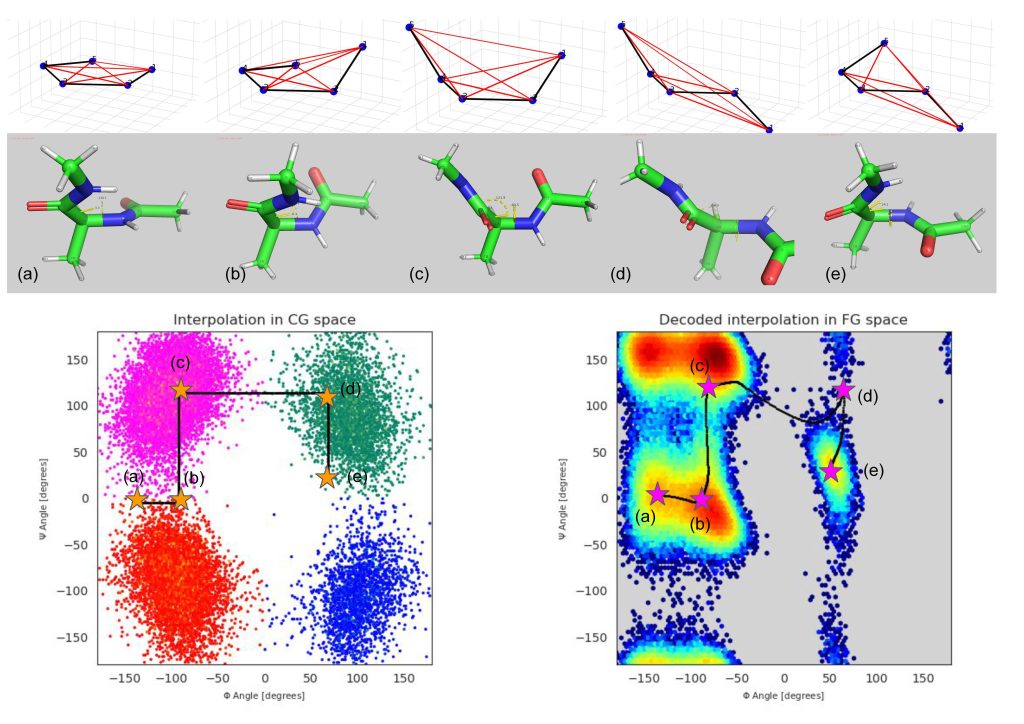
Zhao, Z. (2023). Physically Interpretable Biomolecular Conformation Generation with A Deep Probabilistic Framework (Undergrad Thesis).
(A shout to Minhuan Li and Doeke Hekstra for all the helpful discussions)
TLDR: A hybrid between variational autoencoder (VAE) and normalizing flow (NF) to simultaneously learn a coarse-grained energy function and generate biomolecular conformations.
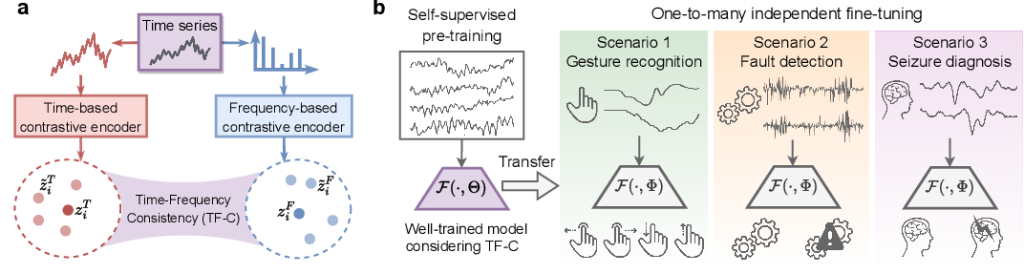
Zhang, X., Zhao, Z., Tsiligkaridis, T., & Zitnik, M. (2022). Self-supervised contrastive pre-training for time series via time-frequency consistency. Advances in Neural Information Processing Systems, 35, 3988-4003.
TLDR: We found that, for time series, pre-training using time- and frequency-based representations from contrastive estimation jointly led to improved performance on downstream tasks.
Research Projects
These are some of the other projects I have worked other than those resulting in the published works shown above.
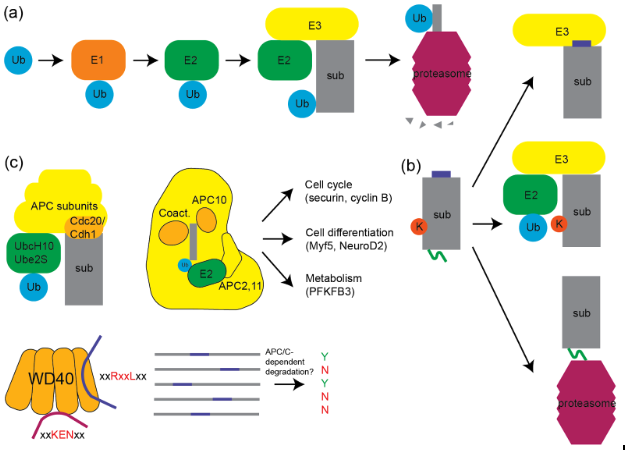
Structure-based modeling and prediction of APC/C substrates (2024)
Shoutout to Ying Lu and Ashwini Jambhekar for their support and to my PQE1 committee: Tim Mitchison, Markus Basan, and Walter Fontana.
TLDR: Predicted substrate specificity for APC/C, important for controlling cell cycle, using coarse-grained molecular dynamics and docking simulations, and achieved superior sensitivity over previous sequence-only models.
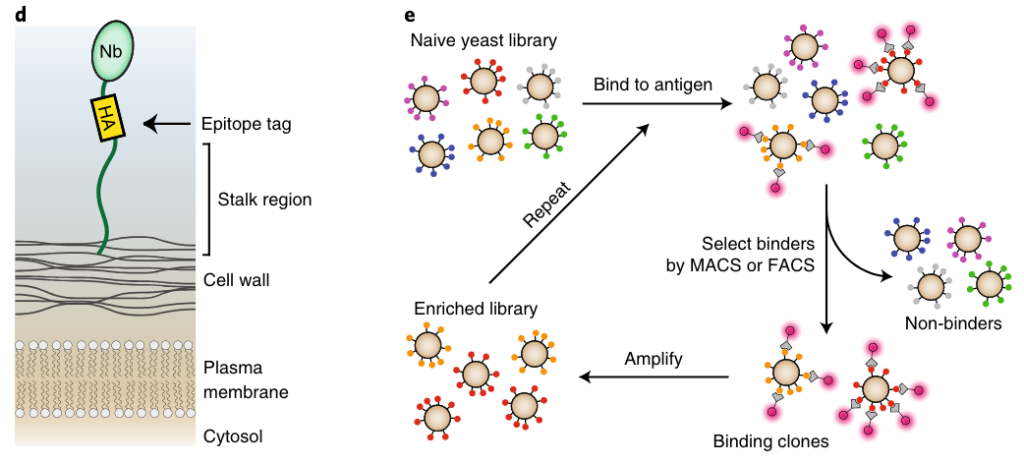
Protein C mAb epitope screening with synthetic nanobody library (2024)
Supervised by Andrew Kruse, mentored by Joe Hurley.
TLDR: With the latest platform developed in the lab, sequencing, and structural modeling, we identified the basis of calcium-dependent epitope affinity against protein C. (Manuscript in preparation)

MTG x Cognitive Psychology (2024-)
TOP SECRET, team reveal early 2025? 😉
TLDR: We want to measure the cognitive differences between Magic: the Gathering players and non-players; we also want to understand why Magic players prefer certain decks, colors, and play styles.
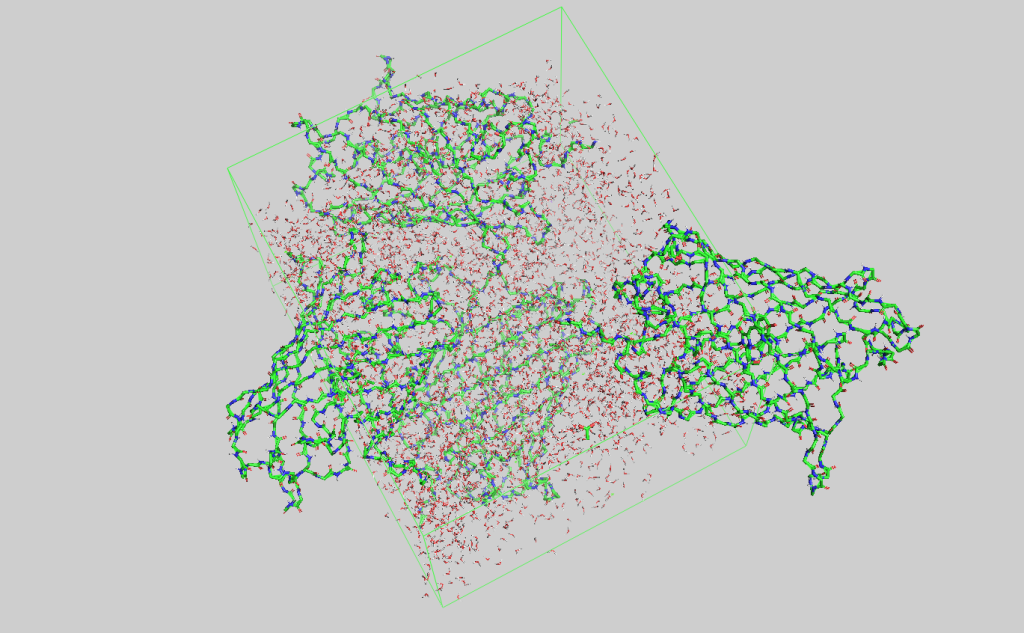
Quantitative comparisons between X-ray crystallography and molecular dynamics simulation (2021-2022)
Supervised by Doeke Hekstra, mentored by Jack Greisman, and funded by HCRP.
TLDR: Corroborated the electron density maps and the difference maps produced from MD simulations of eGFP protein crystal systems with the experimental data. (yeah, the title said it all)
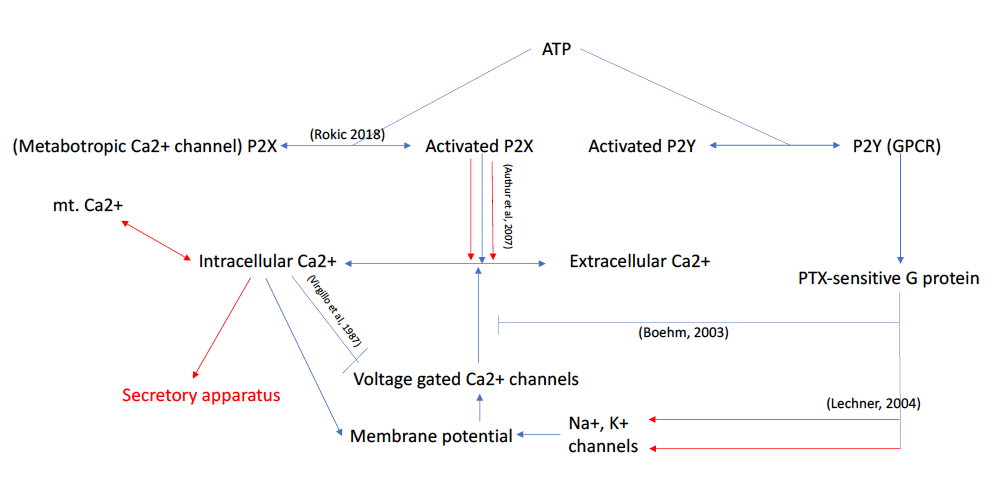
Investigating the learning capacity of single cells (2020-2021)
Supervised by Jeremy Gunawardena, mentored by Rosa Martinez Corral, and funded by PRISE.
TLDR: I developed ODE models based on experimental evidence that suggests some general principles and limitations to how habituation, the simplest type of learning, could happen at the single-cell level.